BioTuring Notebooks
Execute latest methods, algorithms and pipelines, curated and made available as ready-to-use notebooks, coming with instructions and interpretation of example results
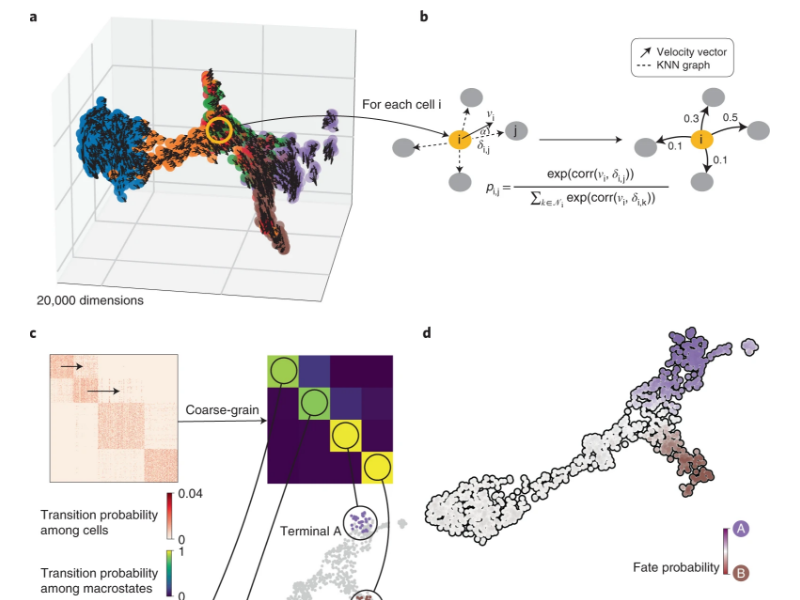
Analyze the dynamics of PPARG-low Tregs precursor populations with Cellrank
We analyze the PPARG-low Tregs precursor population from Chaoran Li and colleagues. The paper reports that lowly-expressed PPARgamma Tregs is indeed transcriptionally heterogeneous. And RNA velocity reveals that its subpopulations exhibit clear transition dynamics to so-called precusor states.
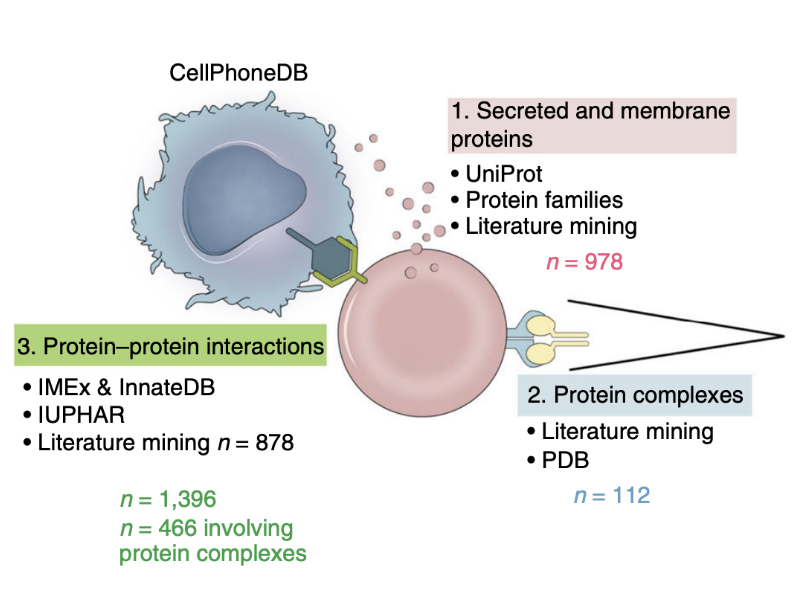
Infer the cell-cell communication with CellphoneDB
We use CellphoneDB v3 to screen for the potential ligand-receptor interactions between Mesenchymal cells (MC), and Hepatic cell (HE) in a late-developed human liver bud. One of the interaction that we found using CellPhoneDB, VEFGA signalling (VEFGA-KDR) was confirmed experimentally in the publication to be a driver of human liver bud development.
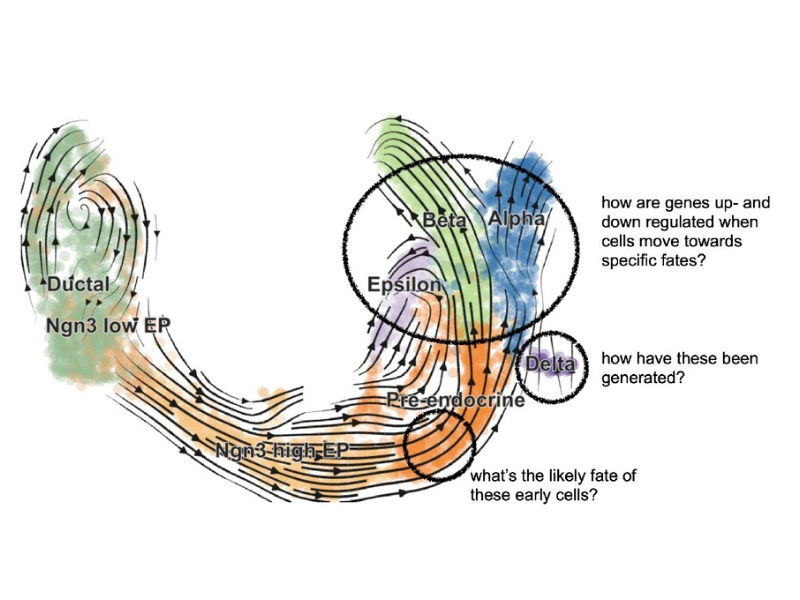
RNA velocity analysis with scVelo
We use scVelo to perform RNA-velocity analysis for a primary tumor sample of patient T71 from the dataset GSE137804: Transcriptomic Characterization of neuroblastoma by Single Cell RNA Sequencing (Donr R. 2020). We identifying two sub-populations of tumor cells in this sample, and use scVelo to predict the progression from one tumor sub-population to another.
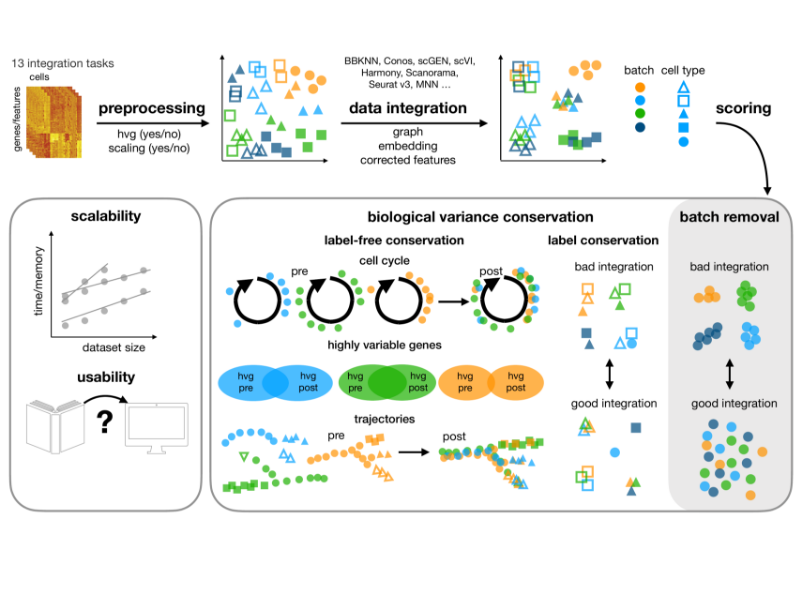
Benchmark single-cell scRNA-Seq data integration with Scib
We use the API from SCIB to evaluation different batch effect removal methods for the human pancreas data that is intergrated from GSE81076, GSE85241, GSE86469, GSE84133, GSE81608.
Gene regulatory inference from scRNA-Seq data using SCENIC
SCENIC package allows users to characterize the single-cell gene regulatory network interference and cluster cell by set of regulons. The example data used in this notebook is the PBMC4k dataset downloaded from 10xGenomics



